Resolving an Insect Outbreak with Next Generation Sequencing
Written on January 1st , 2018 by Bilgenur Baloglu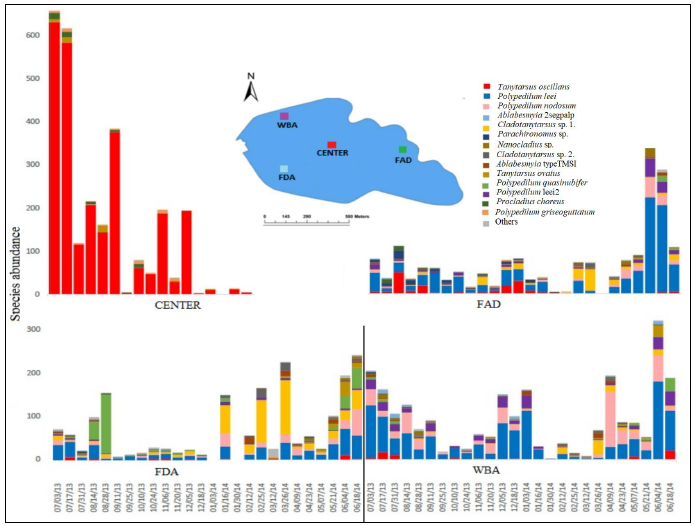
How does NGS solve an insect outbreak problem?
Singapore is an urban city, with several reservoirs storing rain water. One of these reservoirs, Bedok, has experienced several midge swarming events. Chironomid midges are an important family of freshwater invertebrates. Chironomids possess higher species diversity than other macroinvertebrates and have specific habitat requirements that render them useful ecological indicators. Chironomid swarms on the other hand are a health and economic nuisance in urban residential areas around Singapore.
Understanding the life cycle and structure of chironomid communities is essential for identifying the cause of the outbreaks. However, characterizing insect communities is difficult because conventional methods of chironomid species identification based on morphology are costly and laborious, as they usually require the dissection and study of specimens that are mounted on microscopic slides.
Here I used an affordable NGS barcoding approach to study the community structure of chironomid midges in a tropical reservoir. Using COI as barcoding gene, I identified nearly 16,000 specimens. To understand what is causing the outbreaks and locate the larvae responsible for the outbreaks, I studied the temporal and spatial dynamics of chironomid community composition for several sites, dates, sampling methods, and life stages over a period of a year.
With this experiment design, I was able to identify environmental parameters correlated with swarming and determine whether different sampling techniques for midges yield comparable results. I will provide the unpublished manuscript soon.



