Metabarcoding with Nanopore Sequencing
Written on June 6th, 2020 by Bilgenur Baloglu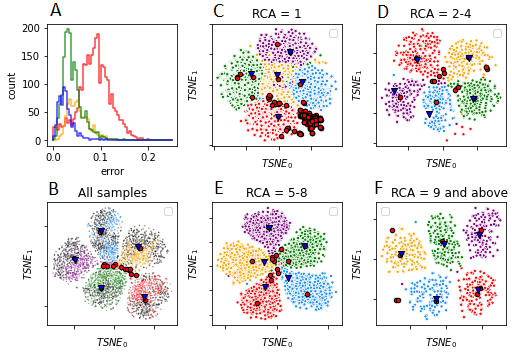
Consensus error correction and density based clustering for metabarcoding
Metabarcoding is a technique used to identify the species composition of a mixed sample using genes such as cytochrome oxidase I - CO1 or ribosomal RNA as taxonomic identifiers. This technique is used in ecological monitoring, outbreak tracking, and disease diagnostics. Most metabarcoding studies to date use establish short read sequencing technologies. Oxford Nanopore MinION, a portable and low-cost sequencing device, is a promising alternative which has enabled researchers to read sequence data in the field. However, a major drawback is the high raw read error rate that can range from 10% to 22%. Our study used consensus error correction and density based clustering to generate highly accurate consensus reads which enabled identification of all 50 species in our mock community. We were able to generate de novo barcodes from consensus reads with comparable accuracy to MiSeq. These results show the feasibility of field based sequencing for biomonitoring and remote diagnostics.


